Experimental Biology

Education
University of the Pacific, Stockton, CA
Doctor of Philosophy, Pharmaceutical and Chemical Sciences
Experimental Skills
Gaining from more than two decades of wet-lab experiences, I have acquired a broad range of technical skills in various domains of the biological science, some of which are listed below:
- Molecular Biology
- Molecular cloning
- DNA/RNA preparations from prokaryotic cells, eukaryotic cells, and animal tissues
- Quantitative RT-PCR (qRT-PCR)
- Northern blot
- Southern blot
- Protein Science
- Protein expression and purification from bacterial, insect and mammalian cells
- Western blot
- Phage display
- Yeast one-hybrid assay, yeast two-hybrid assay, mammalian two-hybrid assay
- Immunoprecipitation assay, co-Immunoprecipitation (co-IP assay)
- ELISA (enzyme-linked immunosorbent assay), cell-based ELISA
- Mammalian Cell Culture
- Generation of CRISPR-induced knockin, knockout and point mutation cell lines
- Maintenance of a multitude of adherent/suspension, and transformed/primary cell lines
- Generation of induced pluripotent stem cells (iPS)
- Generation of neural stem cells
- Immunocytochemistry
- Stable isotope labeling with amino acids assay (SILAC)
- Preparations of lentiviral vectors, adeno-associated virus (AAV) vectors
- Fluorescence microscopy, confocal microscopy
- Flow cytometry
- MTT assay
- Animal Model
- General mouse colony management
- General mouse surgery including tail vein injection, sub-Q (subcutaneous) injection, perfusion and tissue (e.g. hippocampus) harvesting
- Generation of knockout mouse model
- Generation of xenograft and PDX (Patient-Derived Xenograft) mouse models
- PCR- and Southern blot-based genotyping
- Behavioral Testing including locomotor activity, open field behavior, motor coordination (Rotarod), and fear conditioning
- H&E (hematoxylin and eosin) staining
- immunohistochemistry
Current Projects
Cancer Therapeutics
My current projects focus on a rare, nonhereditary cancer Solitary Fibrous Tumor (SFT). SFT is a mesenchymal tumor that demonstrates fibroblastic differentiation and may arise anywhere in the body. In 2013, a defining discovery was made that all SFTs have a version of a hallmark intrachromosomal gene fusion between NAB2 and STAT6 genes on chromosome 12. Yet even though this single gene fusion drives the cancer, the targeted studies, clinical trials, and therapeutic options for SFT are still deficient.
Using CRISPR-based genome editing technology, we have built cell models of SFT which closely match the endogenous NAB2-STAT6 genetic characteristics. In addition, we have established several primary SFT cell lines from resected patient tissues.
Currently, we are adopting a multi-pronged approach to address the therapeutic needs of SFT patients, which include small molecule drug discovery, RNA-based therapeutics (antisense oligonucleotides/ASOs, CRISPR-Cas13d system), and CAR-T cell therapies.
Publications
Genome Engineering
Genetic physical unclonable functions in human cells
A physical unclonable function (PUF) is a physical entity that provides a measurable output that can be used as a unique and irreproducible identifier for the artifact wherein it is embedded. Popularized by the electronics industry, silicon PUFs leverage the inherent physical variations of semiconductor manufacturing to establish intrinsic security primitives for attesting integrated circuits. Owing to the stochastic nature of these variations, photolithographically manufactured silicon PUFs are impossible to reproduce (thus unclonable). Inspired by the success of silicon PUFs, we sought to create the first generation of genetic PUFs in human cells. We demonstrate that these PUFs are robust (i.e., they repeatedly produce the same output), unique (i.e., they do not coincide with any other identically produced PUF), and unclonable (i.e., they are virtually impossible to replicate). Furthermore, we demonstrate that CRISPR-engineered PUFs (CRISPR-PUFs) can serve as a foundational principle for establishing provenance attestation protocols. download
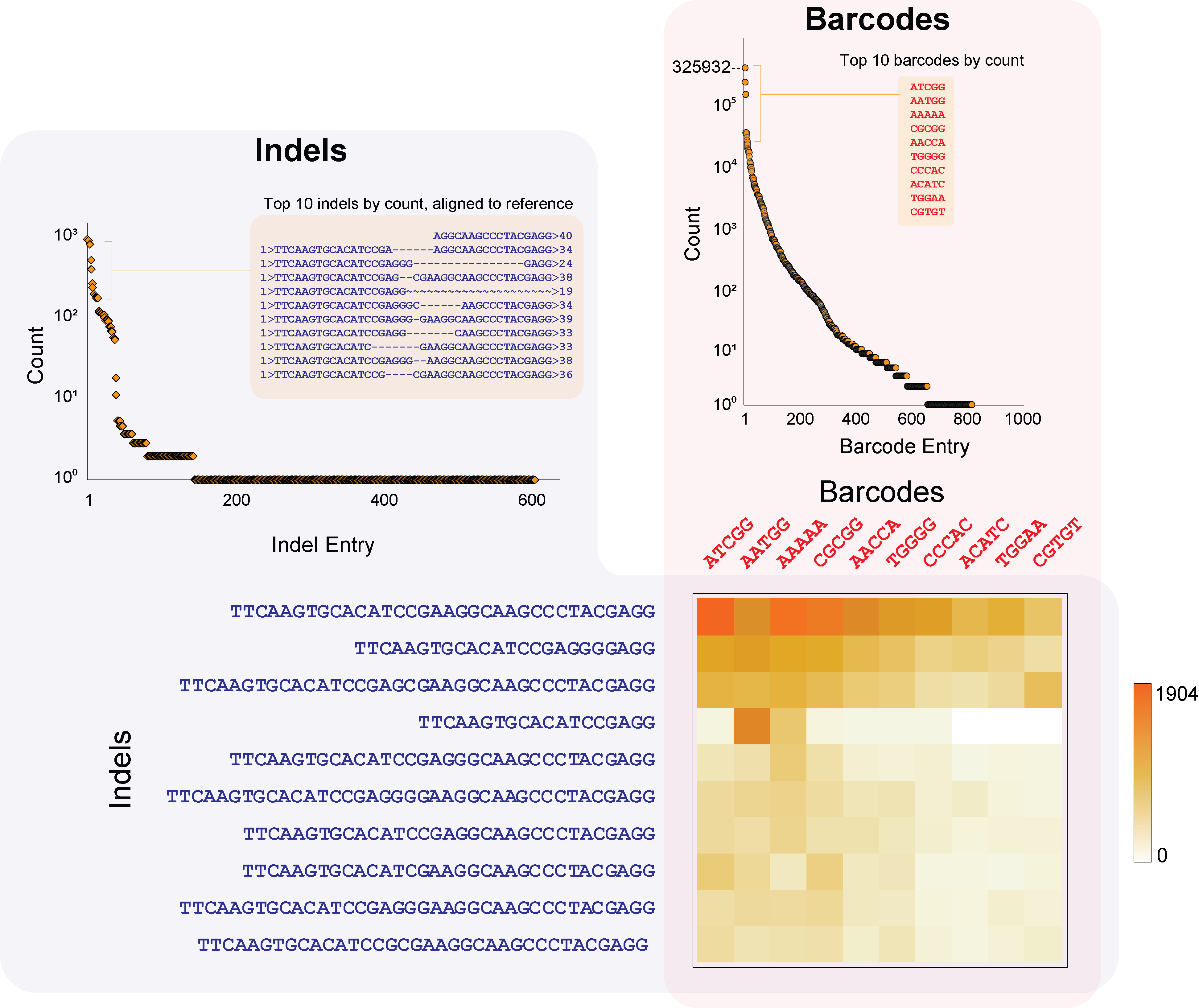
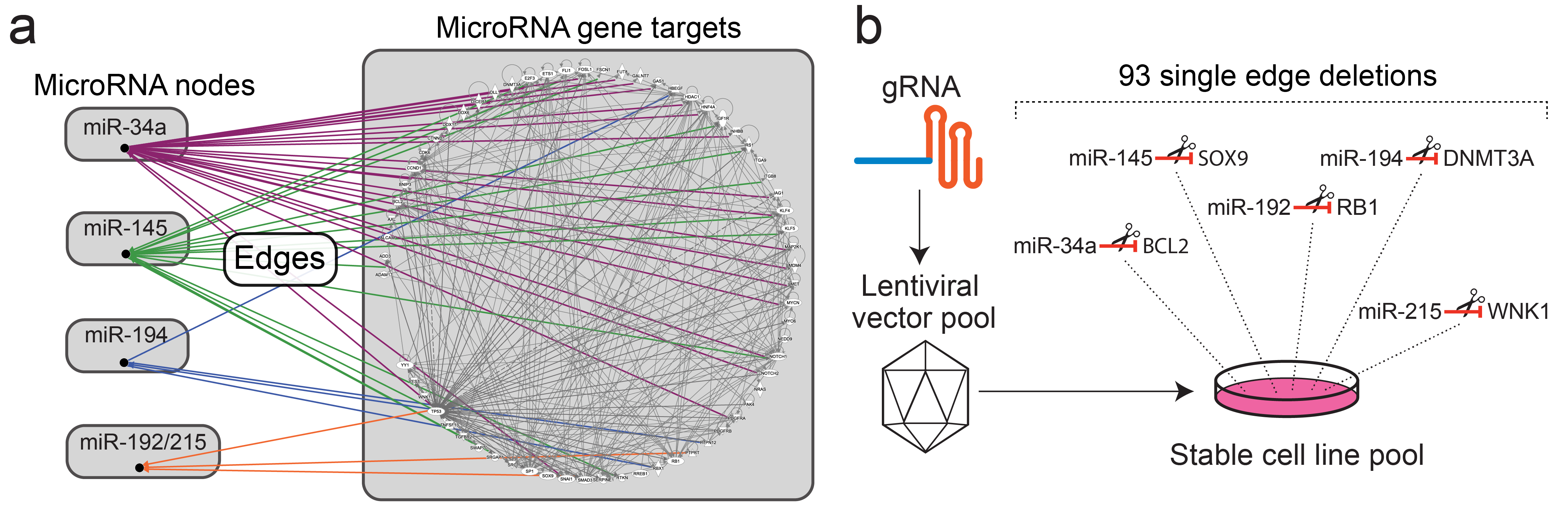
CRISPR-Based Editing Reveals Edge-Specific Effects in Biological Networks
Unraveling the properties of biological networks is central to understanding both normal and disease cellular phenotypes. Networks consist of functional elements (nodes) that form a variety of diverse connections (edges), with each node being a hub for multiple edges. Herein, in contrast to node-centric network perturbation and analysis approaches, we present a high-throughput CRISPR-based methodology for delineating the role of network edges. Ablation of network edges using a library targeting 93 miRNA target sites in 71 genes reveals numerous edges that control, with variable importance, cellular growth and survival under stress. To compare the impact of removing nodes versus edges in a biological network, we dissect a specific p53-microRNA pathway. We show that removal of the miR-34a target site from the anti-apoptotic gene BCL2 desensitizes the cell to ectopic delivery of miR-34a in a p53-dependent manner. In summary, we demonstrate that network edges are critical to the function and stability of biological networks. Our results introduce a novel genetic screening opportunity via edge ablation and highlight a new dimension in biological network analysis. download
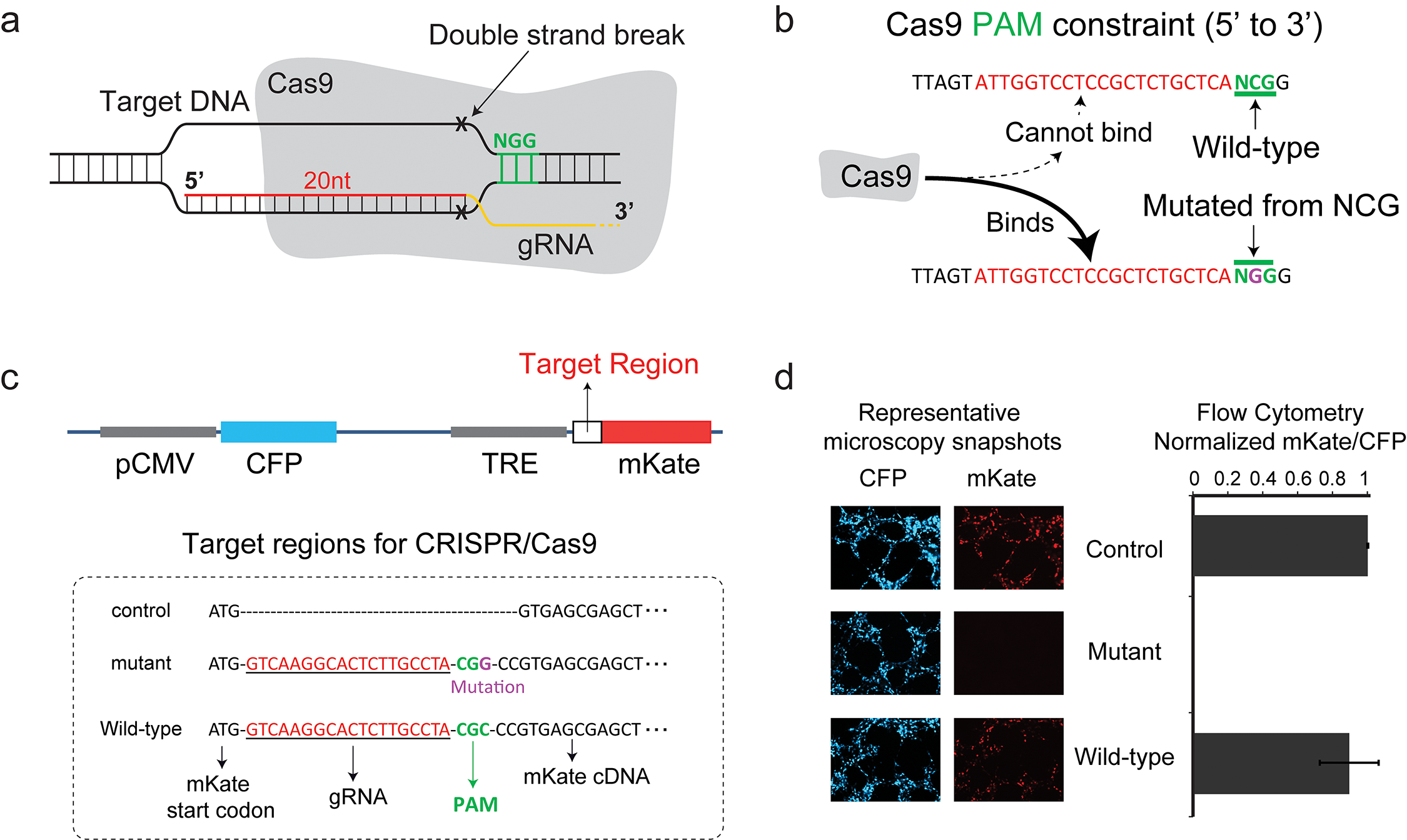
Exploiting the CRISPR/Cas9 PAM Constraint for Single-Nucleotide Resolution Interventions
CRISPR/Cas9 is an enabling RNA-guided technology for genome targeting and engineering. An acute DNA binding constraint of the Cas9 protein is the Protospacer Adjacent Motif (PAM). Here we demonstrate that the PAM requirement can be exploited to specifically target single-nucleotide heterozygous mutations while exerting no aberrant effects on the wild-type alleles. Specifically, we target the heterozygous G13A activating mutation of KRAS in colorectal cancer cells and we show reversal of drug resistance to a MEK small-molecule inhibitor. Our study introduces a new paradigm in genome editing and therapeutic targeting via the use of gRNA to guide Cas9 to a desired protospacer adjacent motif. download
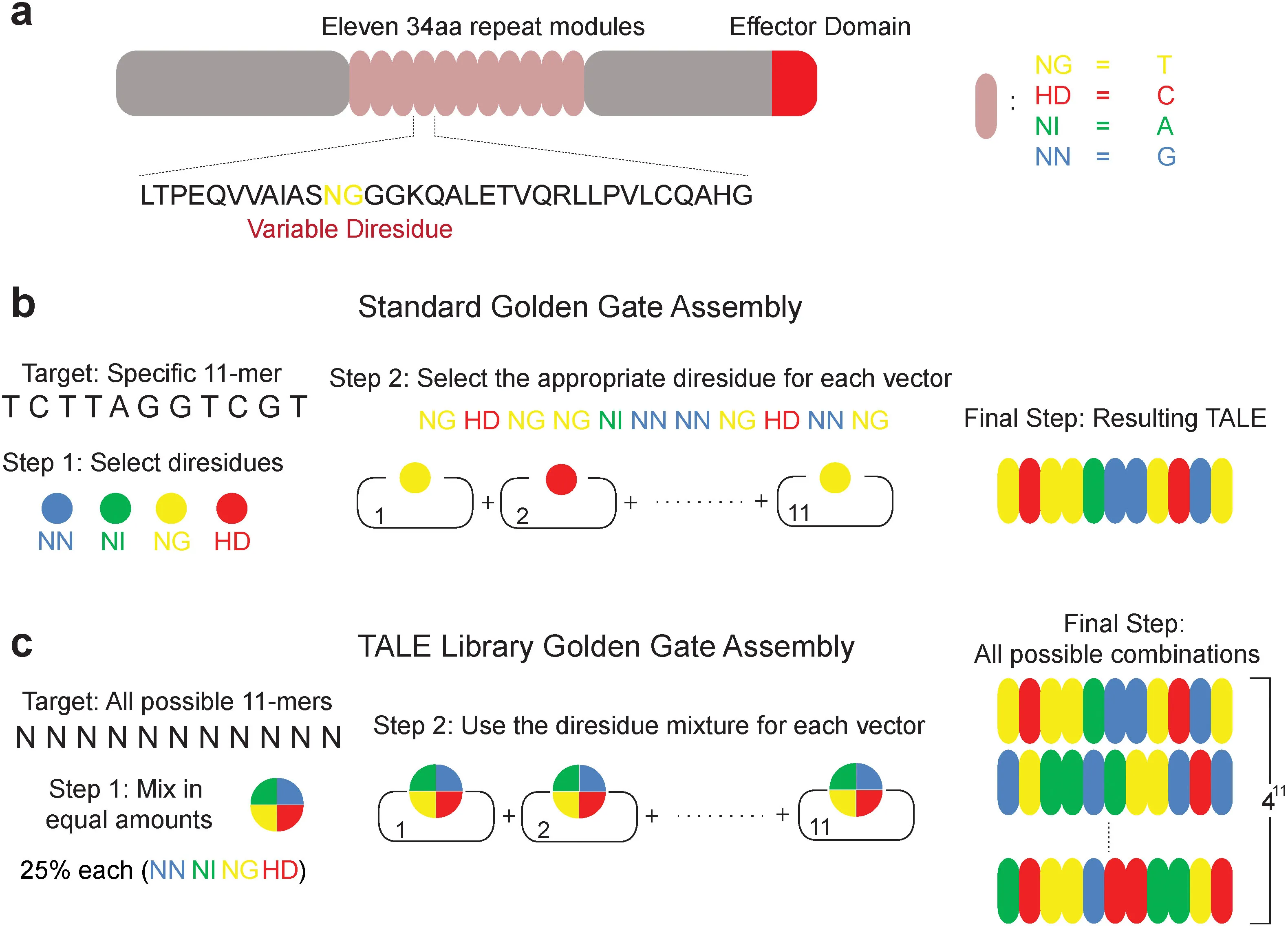
Assembly and Validation of Versatile Transcription Activator-Like Effector Libraries
The ability to perturb individual genes in genome-wide experiments has been instrumental in unraveling cellular and disease properties. Here we introduce, describe the assembly, and demonstrate the use of comprehensive and versatile transcription activator-like effector (TALE) libraries. As a proof of principle, we built an 11-mer library that covers all possible combinations of the nucleotides that determine the TALE-DNA binding specificity. We demonstrate the versatility of the methodology by constructing a constraint library, customized to bind to a known p53 motif. To verify the functionality in assays, we applied the 11-mer library in yeast-one-hybrid screens to discover TALEs that activate human SCN9A and miR-34b respectively. Additionally, we performed a genome-wide screen using the complete 11-mer library to confirm known genes that confer cycloheximide resistance in yeast. Considering the highly modular nature of TALEs and the versatility and ease of constructing these libraries we envision broad implications for high-throughput genomic assays. download
Transcription activator-like effector hybrids for conditional control and rewiring of chromosomal transgene expression
The ability to conditionally rewire pathways in human cells holds great therapeutic potential. Transcription activator-like effectors (TALEs) are a class of naturally occurring specific DNA binding proteins that can be used to introduce targeted genome modifications or control gene expression. Here we present TALE hybrids engineered to respond to endogenous signals and capable of controlling transgenes by applying a predetermined and tunable action at the single-cell level. Specifically, we first demonstrate that combinations of TALEs can be used to modulate the expression of stably integrated genes in kidney cells. We then introduce a general purpose two-hybrid approach that can be customized to regulate the function of any TALE either using effector molecules or a heterodimerization reaction. Finally, we demonstrate the successful interface of TALEs to specific endogenous signals, namely hypoxia signaling and microRNAs, essentially closing the loop between cellular information and chromosomal transgene expression. download
Medical Diagnostics
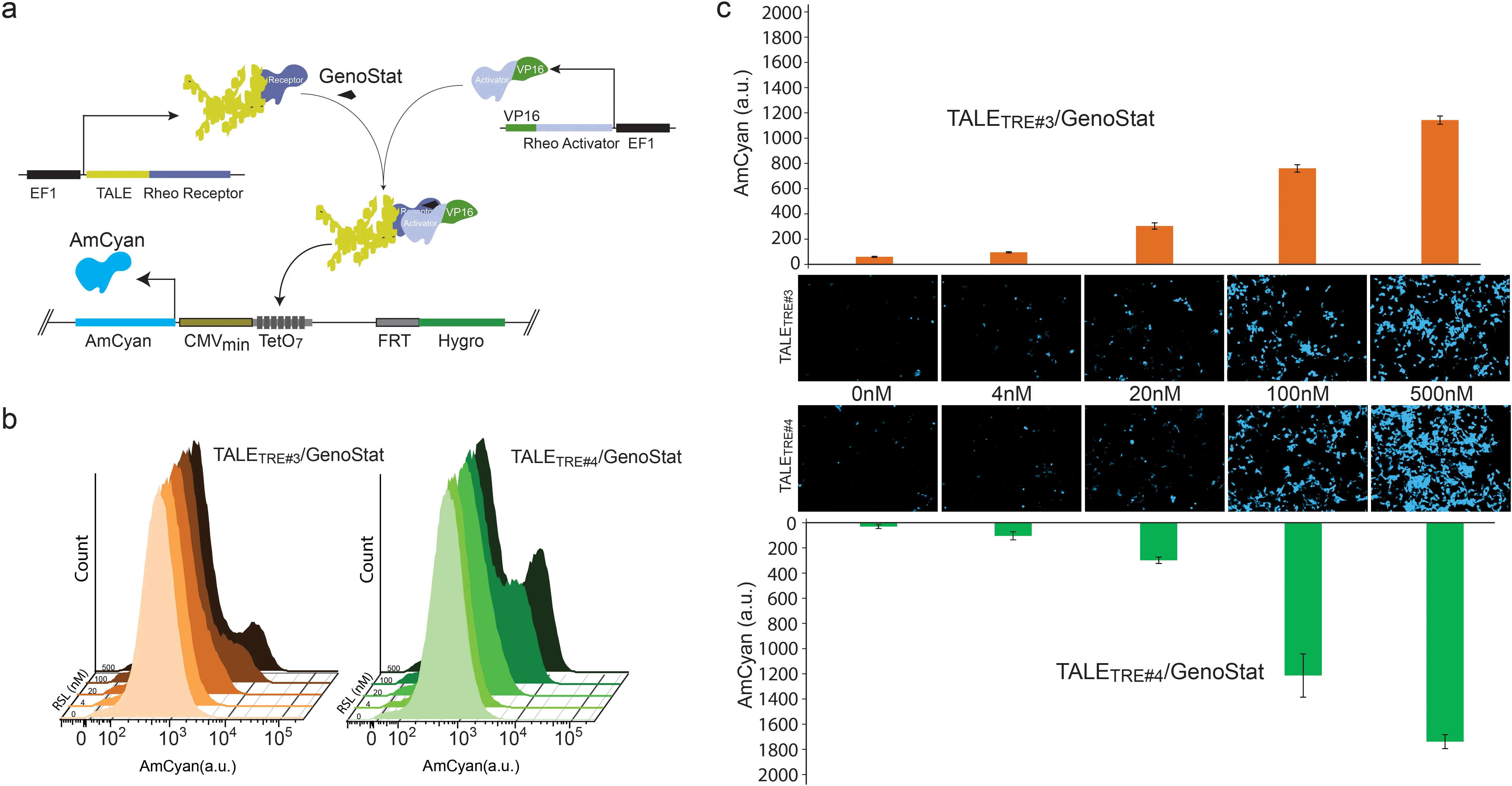
Plasmonic LAMP: Improving the Detection Specificity and Sensitivity for SARS-CoV-2 by Plasmonic Sensing of Isothermally Amplified Nucleic Acids
The ability to detect pathogens specifically and sensitively is critical to combat infectious diseases outbreaks and pandemics. Colorimetric assays involving loop-mediated isothermal amplification (LAMP) provide simple readouts yet suffer from the intrinsic non-template amplification. Herein, a highly specific and sensitive assay relying on plasmonic sensing of LAMP amplicons via DNA hybridization, termed as plasmonic LAMP, is developed for the severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) RNA detection. This work has two important advances. First, gold and silver (Au-Ag) alloy nanoshells are developed as plasmonic sensors that have 4-times stronger extinction in the visible wavelengths and give a 20-times lower detection limit for oligonucleotides over Au counterparts. Second, the integrated method allows cutting the complex LAMP amplicons into short repeats that are amendable for hybridization with oligonucleotide-functionalized Au-Ag nanoshells. In the SARS-CoV-2 RNA detection, plasmonic LAMP takes ≈75 min assay time, achieves a detection limit of 10 copies per reaction, and eliminates the contamination from non-template amplification. It also shows better detection specificity and sensitivity over commercially available LAMP kits due to the additional sequence identification. This work opens a new route for LAMP amplicon detection and provides a method for virus testing at its early representation. download
Neurodegenerative Diseases
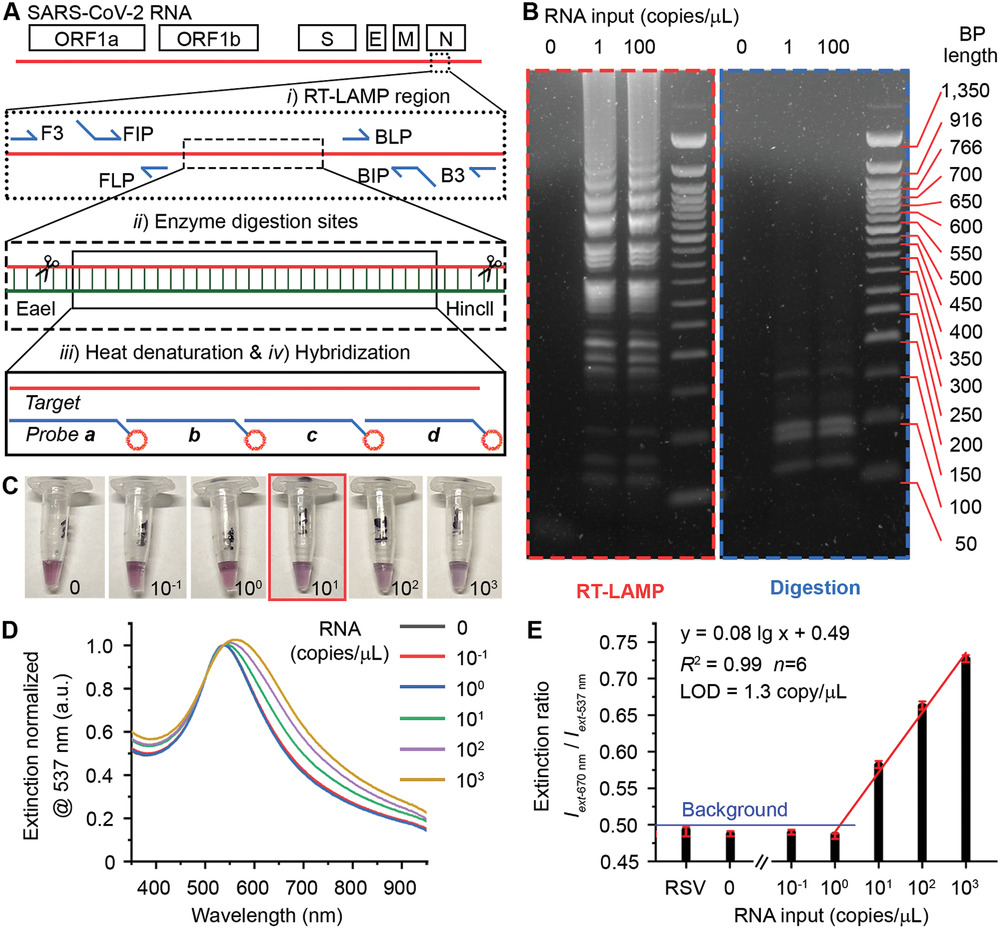
DHHC5 Interacts with PDZ Domain 3 of Post-synaptic Density-95 (PSD-95) Protein and Plays a Role in Learning and Memory
A family of integral membrane proteins containing a signature DHHC motif has been shown to display protein S-acyltransferase activity, modifying cysteine residues in proteins with fatty acids. The physiological roles of these proteins have largely been unexplored. Here we report that mice homozygous for a hypomorphic allele of a previously uncharacterized member, DHHC5, are born at half the expected rate, and survivors show a marked deficit in contextual fear conditioning, an indicator of defective hippocampal-dependent learning. DHHC5 is highly enriched in a post-synaptic density preparation and co-immunoprecipitates with post-synaptic density protein-95 (PSD-95), an interaction that is mediated through binding of the carboxyl terminus of DHHC5 and the PDZ3 domain of PSD-95. Immunohistochemistry demonstrated that DHHC5 is expressed in the CA3 and dentate gyrus in the hippocampus. These findings point to a previously unsuspected role for DHHC5 in post-synaptic function affecting learning and memory. download
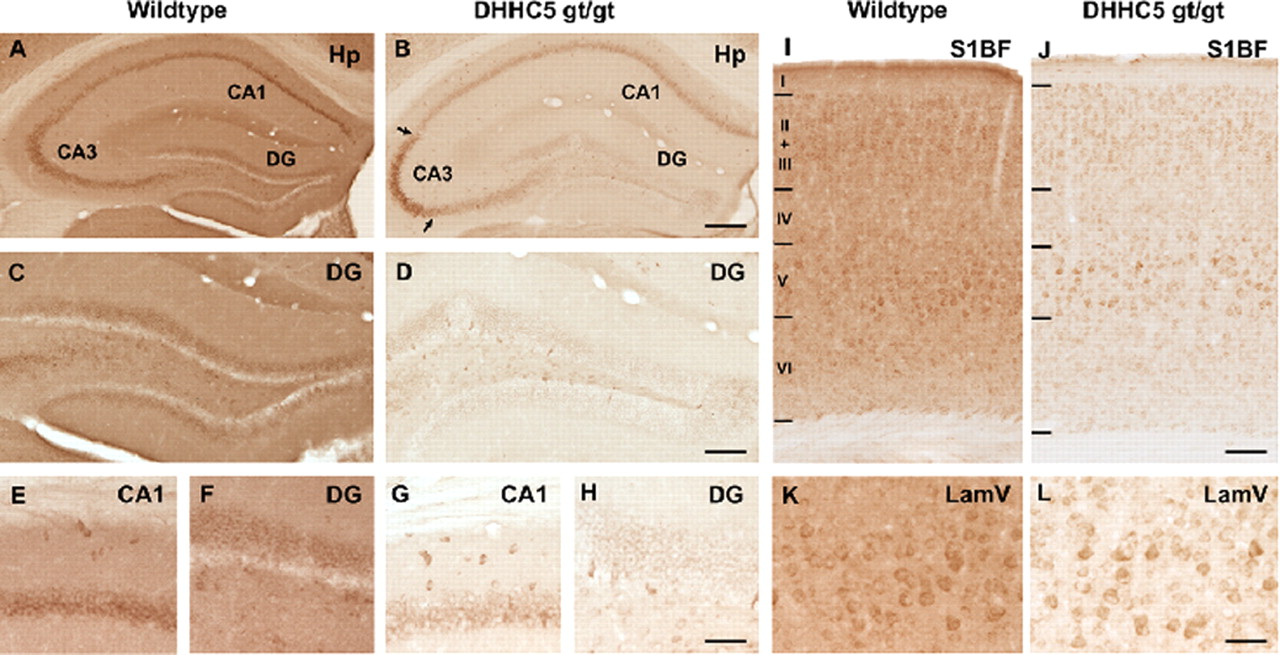
DHHC5 Protein Palmitoylates Flotillin-2 and Is Rapidly Degraded on Induction of Neuronal Differentiation in Cultured Cells
Post-translational palmitoylation of intracellular proteins is mediated by protein palmitoyltransferases belonging to the DHHC family, which share a common catalytic Asp-His-His-Cys (DHHC) motif. Several members have been implicated in neuronal development, neurotransmission, and synaptic plasticity. We previously observed that mice homozygous for a hypomorphic allele of the ZDHHC5 gene are impaired in context- dependent learning and memory. To identify potentially relevant protein substrates of DHHC5, we performed a quantitative proteomic analysis of stable isotope-labeled neuronal stem cell cultures from forebrains of normal and DHHC5-GT (gene-trapped) mice using the bioorthogonal palmitate analog 17-octadecynoic acid. We identified ~300 17-octadecynoic acid-modified and hydroxylamine-sensitive proteins, of which a subset was decreased in abundance in DHHC5-GT cells. Palmitoylation and oligomerization of one of these proteins (flotillin-2) was abolished in DHHC5-GT neuronal stem cells. In COS-1 cells, overexpression of DHHC5 markedly stimulated the palmitoylation of flotillin-2, strongly suggesting a direct enzyme-substrate relationship. Serendipitously, we found that down-regulation of DHHC5 was triggered within minutes following growth factor withdrawal from normal neural stem cells, a maneuver that is used to induce neural differentiation in culture. The effect was reversible for up to 4 h, and degradation was partially prevented by inhibitors of ubiquitin-mediated proteolysis. These findings suggest that protein palmitoylation can be regulated through changes in DHHC PAT levels in response to differentiation signals. download
Signaling Pathway
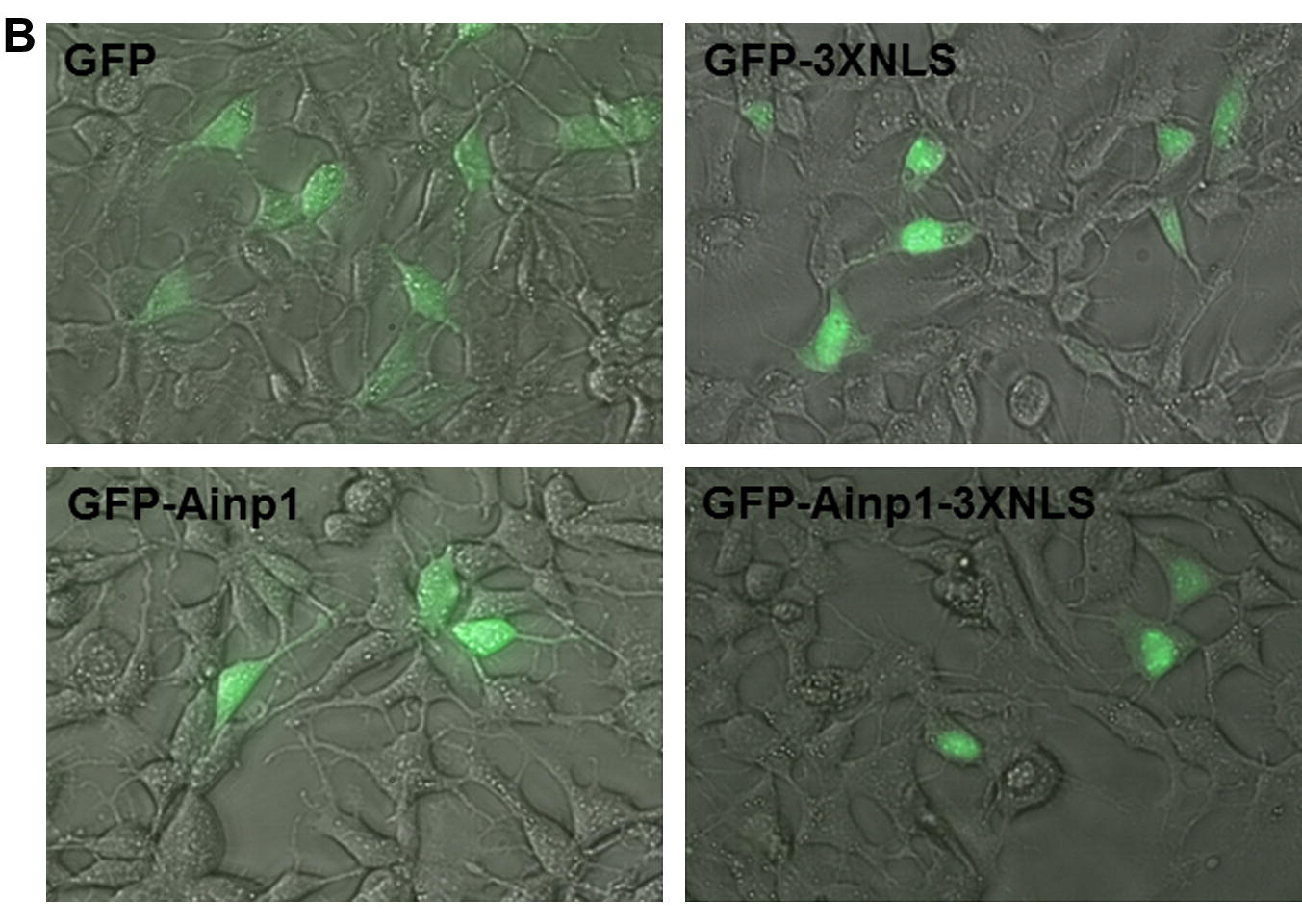
Suppression of the hypoxia inducible factor-1 function by redistributing the aryl hydrocarbon receptor nuclear translocator from nucleus to cytoplasm
The aryl hydrocarbon receptor nuclear translocator (ARNT) heterodimerizes with hypoxia inducible factor-1α (HIF-1α), followed by upregulation of genes that are essential for carcinogenesis. We utilized a novel peptide (Ainp1) to address whether the HIF-1α signaling could be suppressed by an ARNT-mediated mechanism. Ainp1 suppresses the HIF-1α-dependent luciferase expression in Hep3B cells and this suppression can be reversed by ARNT. Ainp1 reduces the interaction between ARNT and HIF-1α, suppresses the formation of the HIF-1 gel shift complex, and suppresses the ARNT recruitment to the vegf promoter. These effects are partly mediated by redistribution of the nuclear ARNT contents to the cytoplasm. download
The aryl hydrocarbon receptor nuclear translocator-interacting protein 2 suppresses the estrogen receptor signaling via an Arnt-dependent mechanism
We explored whether modulation of the estrogen receptor (ER) signaling is possible through an aryl hydrocarbon receptor nuclear translocator (Arnt)-dependent mechanism. We utilized the Arnt-interacting protein 2 (Ainp2) to examine whether the presence of Ainp2 in MCF-7 cells would interfere with the Arnt-mediated ER signaling. We found that Arnt increased the 17 beta-estradiol (E2)-dependent luciferase activity and Ainp2 significantly suppressed this Arnt-mediated luciferase activity. Ainp2 significantly suppressed 25% of the E2- and Arnt-dependent upregulation of the GREB1 message. No suppression of the ER target gene expression by Ainp2 was detected in Arnt-knockdown MCF-7 cells and in Arnt-independent ER signaling. Although Ainp2 did not interact with ER alpha and ER beta, it suppressed the ER alpha::Arnt interaction and reduced the E2- driven recruitment of Arnt to the GREB1 promoter. We concluded that Ainp2 suppresses the ER signaling by not allowing Arnt to participate in the ER-dependent, Arnt-mediated activation of gene transcription. download
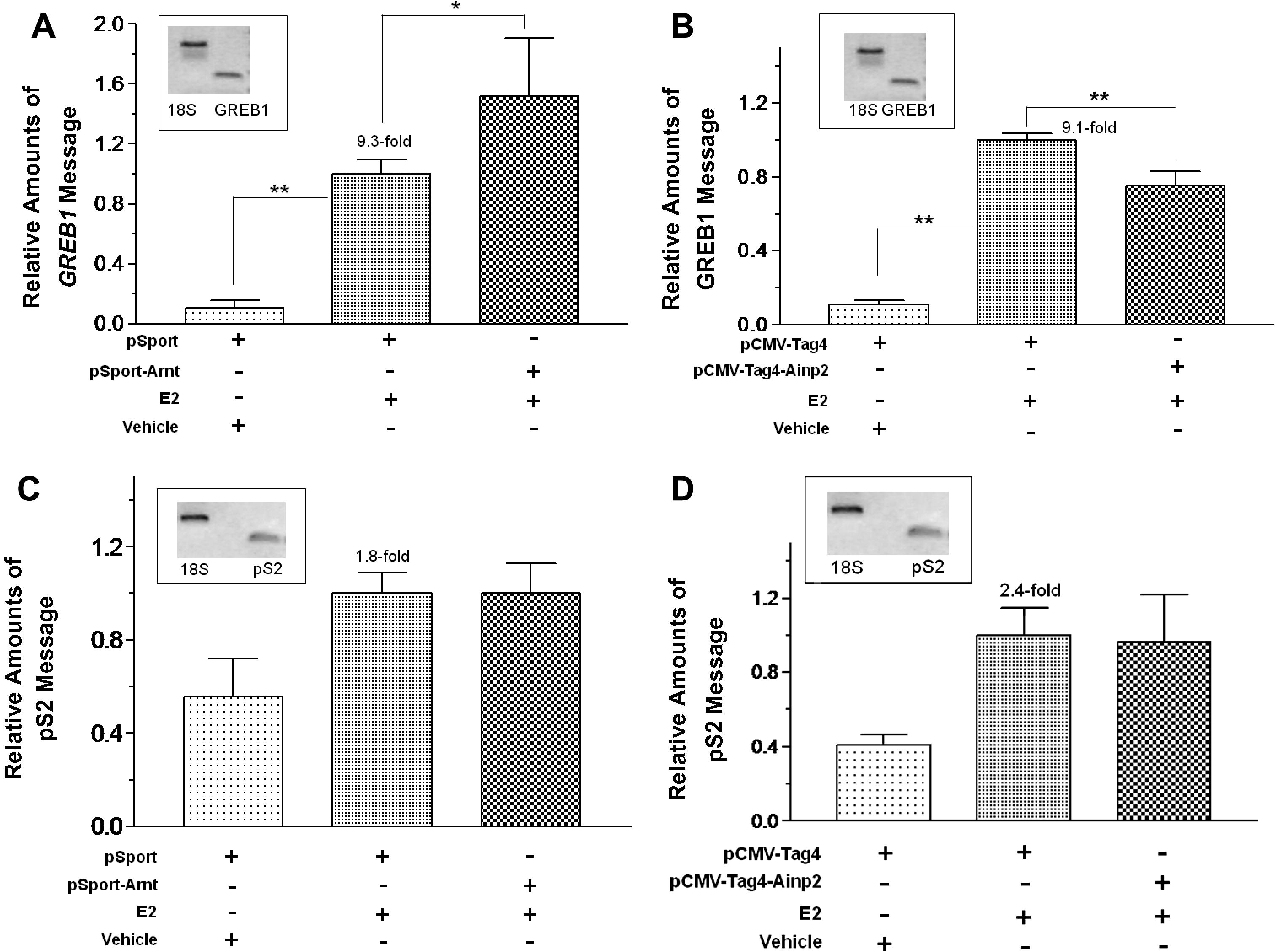
A novel Arnt-interacting protein Ainp2 enhances the aryl hydrocarbon receptor signalling
In an effort to better understand the Ah receptor nuclear translocator (Arnt)-dependent signaling mechanisms, we employed a phage display system to identify Arnt-interacting peptides. Human liver cDNA library was utilized to screen for Arnt- interacting peptides using an Arnt construct fused to thioredoxin (TH-ArntCΔ418). Two clones, namely Ainp1 and Ainp2 (Arnt-interacting peptide), were identified and subsequently Ainp2 was further characterized. Ainp2 interacts with TH-ArntCΔ418 in the GST pull-down and mammalian two-hybrid assays. Northern blot results revealed that Ainp2 is predominantly expressed in human liver. The putative full-length Ainp2 cDNA sequence was subsequently cloned using RACE PCR. Endogenous expression of Ainp2 was found in Jurkat cells at the mRNA and protein levels. Results from the transient transfection studies using a DRE-driven reporter plasmid and the real-time QPCR experiments examining the endogenous CYP1A1 expression showed that Ainp2 enhances the 3-methylchloranthrene-induced activity in HepG2 cells, suggesting that Ainp2 plays a role in the Arnt-dependent function. download
